Constructing a machine: controlled movements
Abstract
We would now like to turn our attention to the editor and take the first step in this tutorial to compose our own machine. For this purpose, we will build a machine that accelerates alternately forward and backward. It will familiarize us with two cell functions: One for calculating with memory contents (computer cell function) and one for accelerating (propulsion cell function).
What you will see
1. Building a cell skeleton
We want to start by assembling a small cell cluster as a foundation. To do this, we create a new simulation and set the simulation parameter radiation → factor to 0. This ensures that our construction will not be destroyed by radiation emission over time. We then zoom in to 32x, such that that the editor including an additional toolbar opens automatically. There, we create six cells by clicking 6 times on the first button of the editor toolbar (alternatively in the menu Entity → New cell). The result could then look like this:
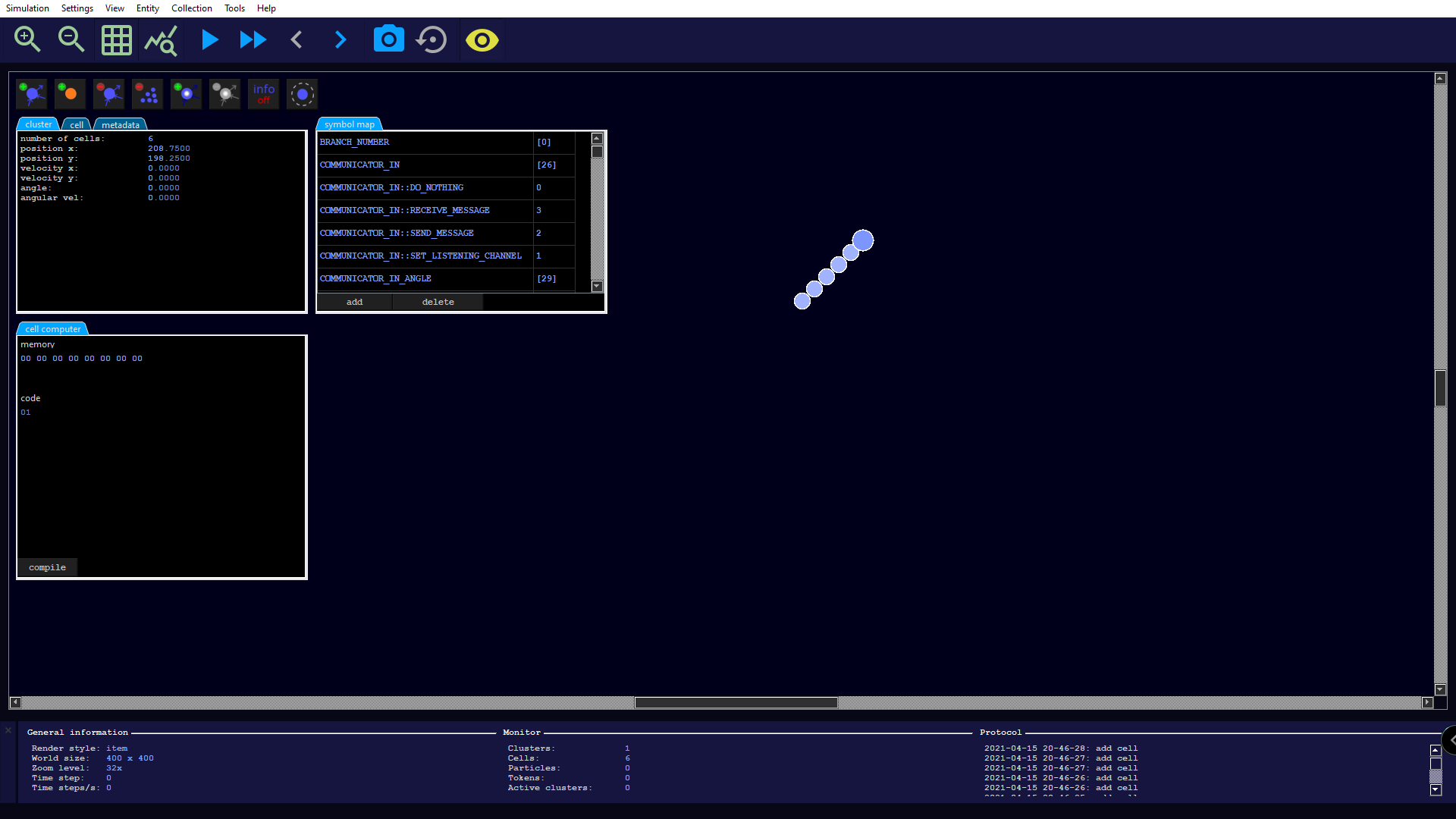
We arrange the cells into a small circle using drag and drop, making sure that they still remain connected to each other to form a single cell cluster. As a further visual help we activate the cell information by pressing the button labeled Info off (alternatively we can also reach it in the menu under View → Cell info).

2. Setting token branch numbers
The next step is to define an order on the cell cluster for triggering the cell functions by tokens. For this purpose a so-called branch number is assigned to each cell. Therewith one can specify a direction between two connected cells as follows:
- A cell X points towards a connected cell Y if Y has a branch number increased by one compared to X.
- If the branch number of a cell X has the highest possible value (configurable by a simulation parameter) and a connected cell Y has the value 0, then there is also a directed connection from X to Y.
- In all other cases, there is no directed connection between two cells.
The idea behind this is that if there is a token on a cell, the token knows on which connected cells it should move at the next time step. By default there are 6 possible branch numbers ranging from 0 to 5.
We now assign clockwise ascending branch numbers to the individual cells of our circular cluster. Therefore, after selecting a cell, we can set the branch number in the cell tab (top left). This value is additionally displayed directly on the cell. Please note that we need to confirm each number entry with enter. If everything worked correctly, our little machine should look like this:
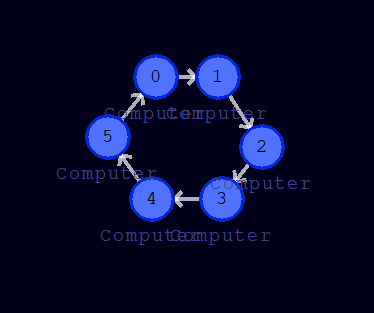
We also see that the directions of movement of the tokens given by the branch number are displayed. Due to these inputs, we have created a directed graph on which our tokens can rotate eternally in the circular structure (at least as long as there are no disturbing influences from outside).
3. Setting cell functions, token and metadata
The plan is as follows: Of the six cells, we will equip two with propulsion functions. One will be used to alternately accelerate forward and backward, while the other one will dampen the angular velocity that may occur due to internal or external forces. We click on the cell with the branch number 5 and then select the cell function Propulsion in the cell tab. We also switch to the metadata tab and select a different color, e.g. green. We do the same with cell 2. The remaining cells are left with the default cell function Computer. We should now see the following:

In order for the cell functions to be triggered, we still need to create a token. The token should initially be placed on a cell with branch number 0 (this restriction can be bypassed, but that will not discussed here). The token is visually represented by a small white circle.
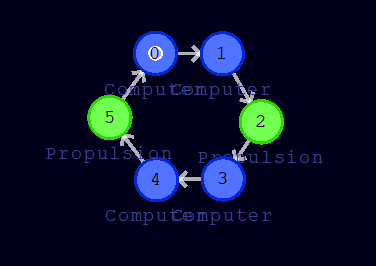
4. Cell program for damping angular velocity
Our previous design would not be able to do anything yet. What is missing is to set the appropriate control commands for the propulsion cells. In general, as soon as a token passes a cell, the token memory is used to extract the control commands for the cell functions. Hence, we need to set the appropriate control commands in the token memory via a little program in the computer cell functions. The computer cell functions are programmed with a kind of machine language, which are compiled from an assembler-like language.
As a first task, we will reprogram the cell with branch number 1 to write a command in the token memory that will cause the propulsion cell to dampen the angular velocity. The program on this cell will be executed when the token is one time step away from a propulsion cell.
As soon as we click on the cell with branch number 1, a tab opens for entering the program code:
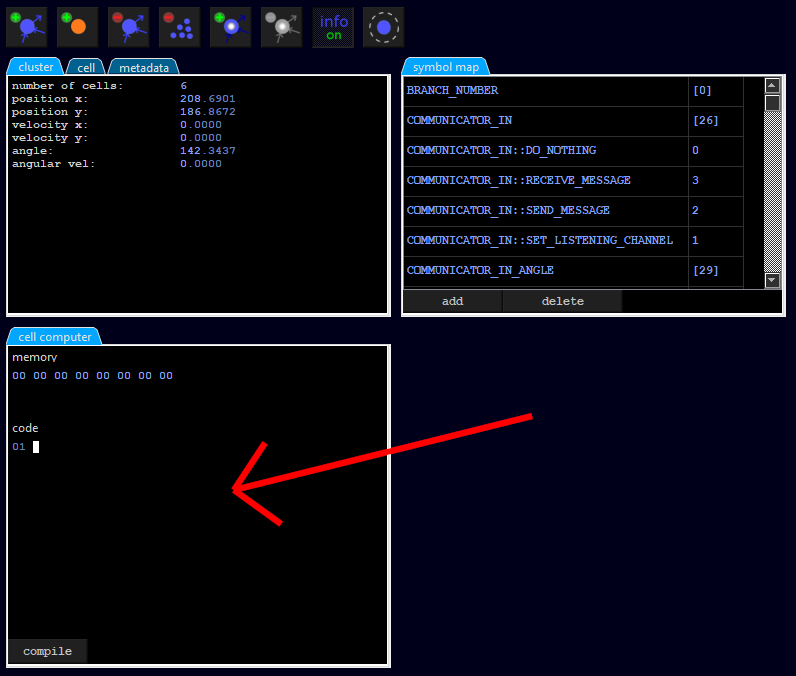
We enter the following line there:
mov PROP_IN, PROP_IN::DAMP_ROTATION
This instruction copies into the memory location (of the token) named PROP_IN a value named PROP_IN::DAMP_ROTATION. Both PROP_IN and PROP_IN::DAMP_ROTATION are symbols defined in a dictionary called symbol map (see the tab symbol map). For example, PROP_IN is defined as [8], which simply means the memory content from the 9th byte of the token (counting from 0). Furthermore, PROP_IN::DAMP_ROTATION is defined as the constant 6. The symbols are just there to write more readable code, because above command is more understandable than
mov [8], 6
After entering the line, we click on the compile button so that the source code is translated into machine code and transferred to the cell. If a compile error occurs, this is indicated by a short message specifying the line number. In our case, everything should be OK:
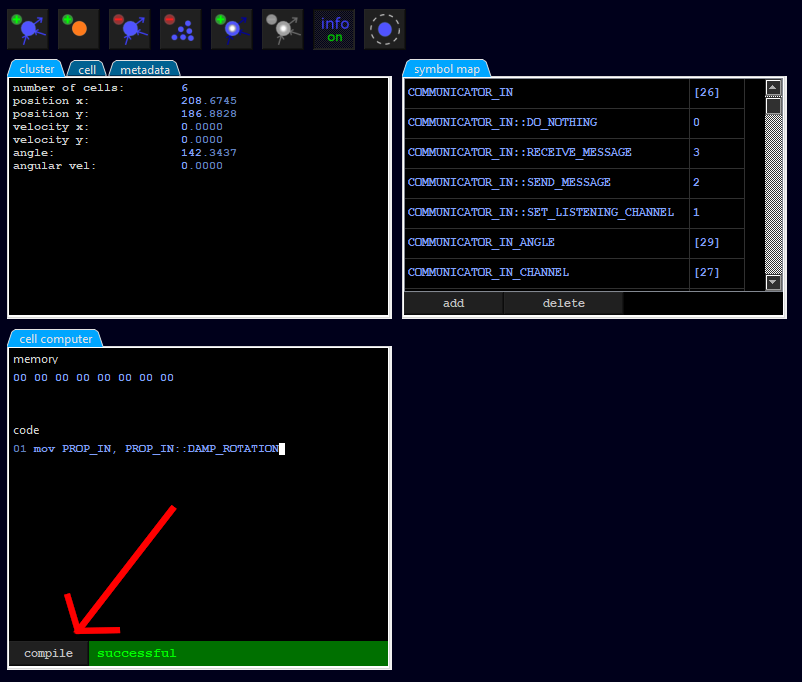
One more remark: We see that a memory content is displayed above the source code. This is not the memory of the token (the token is currently placed on cell 0), but the memory of the cell. By default the token memory is 256 bytes and the cell memory is 8 bytes large.
5. Cell program for accelerating forward and backward
At last we have to implement the most complicated part: The control commands for the forward and backward movements. In order to do this, we insert the following program code to cell 4, which is executed by the token before it reaches the propulsion cell 5:
01: mov PROP_IN, PROP_IN::TOWARD_CENTER
02: if i < 32
03: mov PROP_IN, PROP_IN::FROM_CENTER
04: endif
05: if i > 95
06: mov PROP_IN, PROP_IN::FROM_CENTER
07: endif
08: mov PROP_IN_POWER, 200
09: add i,1
10: if i = 128
11: mov i, 0
12: endif
Let us go through the code line by line:
- Line 1: In the first line, we set the command that causes the propulsion cell to generate thrust in the direction of the center of mass.
- Line 2-4: However, if the variable i (is predefined as a symbol and initialized as 0) is smaller than 32, then the thrust should take place exactly in the opposite direction.
- Line 5-7: The same should happen if i is greater than or equal to 96=32*3.
- Line 8: Here we specify the power of the thrust. The semantic is as follows: 0 (no thrust) and 255 (maximum thrust).
- Line 9-12: We increase the value of i by 1. If the value exceeds 128=32*4, we reset it to 0.
The way of working should become clear now: The token rotates around our cell cluster and after each rotation our program is executed. A full acceleration interval comprises 64 executions. At the beginning i has the value 0 and therefore performs only an acceleration for half an interval (32 executions). After that, we accelerate for a full interval (64 executions) in the opposite direction. Half an interval (32 executions) of this is needed to decelerate the machine back to 0 and another half interval is used to accelerate further in the opposite direction. After that the acceleration turns around again and the procedure starts from the beginning.
After inserting the program code you have to click compile again and our machine is ready!
For testing purposes it is useful to save the machine and to limit the execution speed to 100 time steps per second (in the menu under Simulation → Restrict time steps per seconds). During the simulation it is noticeable that a particle stream is emitted only in one direction. This is due to the fact that the energy particles in the other direction collide with their own cell cluster and are absorbed.
Created with the Personal Edition of HelpNDoc: Create HTML Help, DOC, PDF and print manuals from 1 single source